Tutorial
Creating the root of the tree
A tree is represented by a Node. First, let’s create the root:
from littletree import Node
tree = Node({"population": 7.909}, identifier="World")
All nodes have the following attributes:
Attribute |
Description |
|---|---|
|
Identifier of the node. Must be unique among siblings. |
|
Parent node. |
|
Child nodes. |
|
Dictionary for storing arbitrary data. |
Adding nodes
You can add child nodes via assignment:
tree["Asia"] = Node({"population": 4.694})
tree["Africa"] = Node({"population": 1.393})
Or by providing children or a parent in the constructor:
# Adding children during creation
tree = Node(
{"population": 7.909},
identifier="World",
children={
"Asia": Node({"population": 4.694}),
"Africa": Node({"population": 1.393})
}
)
# Assigning a parent
Node({"population": 1.029}, identifier="America", parent=tree)
Restrictions
A node cannot have multiple parents:
tree1 = Node()
tree2 = Node()
child = Node()
tree1["child"] = child # OK
tree2["child"] = child # Raises DuplicateParentError
Use detach() or copy() to reuse nodes:
tree2["child"] = child.detach() # Move node
tree2["child"] = child.copy() # Copy node
Sibling identifiers must be unique:
tree = Node()
child1 = Node(identifier="name", parent=tree)
child2 = Node(identifier="name", parent=tree) # DuplicateChildError
child2 = Node(identifier="different_name", parent=tree) # OK
Cycles are not allowed:
tree = Node()
tree["child"] = tree # LoopError
Bulk adding
update() allows adding multiple children efficiently:
tree.update({
"Asia": Node({"population": 4.694}),
"Africa": Node({"population": 1.393}),
})
You can copy or detach nodes from another tree:
tree1 = Node(identifier="tree1")
tree2 = Node(identifier="tree2")
tree1["Asia"] = Node({"population": 4.694})
# Copy node from tree1 to tree2
tree2.update([tree1["Asia"]], mode="copy")
# Move node from tree1 to tree2
tree2.update([tree1["Asia"]], mode="detach")
Iteration
Common iterators:
Iterator |
Description |
|---|---|
|
Iterate over children |
|
Iterate from root to self |
|
Iterate over all nodes |
|
Iterate over ancestors |
|
Iterate over descendants |
|
Iterate over siblings |
|
Iterate over multiple trees together |
|
Iterate from one node to another |
The nodes can be iterated in a specific order.
Along with each node, an item is also yielded.
You can access item.depth to get the node’s depth in the tree, and item.index to get its position among siblings at that level.
for node, item in tree.nodes.preorder():
...
for node, item in tree.nodes.postorder():
...
for node, item in tree.nodes.levelorder():
...
Optional keep argument allows filtering nodes and their descendants:
# Limit iteration to depth 2
for node, item in tree.nodes.preorder(keep=MaxDepth(2)):
...
Path operations
Get the path to a node:
str(tree["Europe"].path) # "/World/Europe"
Access nodes using paths:
lisbon = tree["Europe"]["Portugal"]["Lisbon"] # Bracket style
lisbon = tree.path(["Europe", "Portugal", "Lisbon"])
lisbon = tree.path("Europe/Portugal/Lisbon")
Create nodes along a path if they don’t exist:
lisbon = tree.path.create(["Europe", "Portugal", "Lisbon"])
lisbon = tree.path.create("Europe/Portugal/Lisbon")
Search using glob patterns:
lisbon_nodes = tree.path.glob("**/Lisbon")
Miscellaneous tree operations
Height and breadth of the tree:
height = tree.levels.count() - 1
breadth = tree.leaves.count()
Depth of a node:
depth = node.ancestors.count()
Path from one node to another:
madrid = tree.path.create("Europe/Spain/Madrid")
madrid_to_lisbon = list(madrid.to(lisbon))
Distance between nodes (edges):
distance = madrid.to(lisbon).edges.count()
Lowest common ancestor:
europe = madrid.to(lisbon).lca
Exporting and serialization
Function |
Description |
|---|---|
|
Pretty print the tree |
|
Plot with matplotlib |
|
Export to dot, mermaid, or LaTeX |
|
Requires Graphviz or Mermaid |
|
Convert to/from JSON-like formats |
|
Convert to/from path lists |
|
Convert to/from parent-child lists |
|
Convert to/from Newick format |
|
Convert to/from networkx graphs |
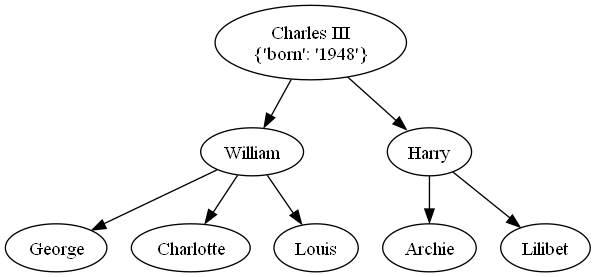
Example using Newick format:
family_newick = "((George, Charlotte, Louis)William,(Archie, Lilibet)Harry)'Charles III'[&&NHX:born=1948]"
family_tree = Node.from_newick(family_newick)
family_tree.show(style="round-arrow")
family_tree.to_pillow().show() # Requires Graphviz and Pillow
family_tree.plot() # Requires matplotlib
Charles III
{'born': '1948'}
├→ William
│ ├→ George
│ ├→ Charlotte
│ ╰→ Louis
╰→ Harry
├→ Archie
╰→ Lilibet